YALES2BIO public page
Contents
Motivation
YALES2BIO aims at the solving of the equations of fluid dynamics in the context of biological flows. The main application is cardiovoscular flows, involving microscale (at the level of cells) and macro-scale applications (blood flow in the heart).
YALES2BIO was developed from 2010 to 2013 by S. Mendez and C. Chnafa and is maintained since 2012 by S. Mendezat I3M, together with the YALES2 devbelopers at CORIA.
Solvers
YALES2BIO is based on YALES2. In addition to the solvers implemented in YALES2, YALES2BIO has several specific solvers.
- Red blood cell solver (RBC)
- Electrostatics solver (ESS)
- ALE solver, with specific routines for heart (ALE)
All the following features are inherited from the YALES2 kernel:
Models
- Turbulence (Large-Eddy Simulation)
- Constant Smagorinsky
- Localized dynamic Smagorinsky
- WALE
- SIGMA
Numerics
- Spatial: 2nd- and 4th-order finite-volume schemes
- Temporal:
- 4th-order explicit time integration (RK4 and TFV4A) of convective terms
- explicit and implicit diffusion and source terms
- Stabilization: Cook & Cabot 4th-order artificial viscosity
- Linear solvers:
- PCG
- BICGSTAB, BICGSTAB2, BICGSTAB(2)
- Deflated PCG
- Deflated BICGSTAB(2)
- Residual recycling
Data Structures
- 1D, 2D, 3D unstructured solver
- Full dual decomposition based on METIS
- Data registration (int, real, char, node, elem, face, pair, scalar, vector, tensor)
- Optimized non-blocking MPI communications
- Parallel load balancing
- Automatic reconnection of periodic boundaries
- Automatic homogeneous mesh refinement
- IO formats: Gambit (Fluent), Ensight, prepartionned HDF5 (XDMF) with compression
- Cartesian mesh generator
- Partitioned mesh support for HDF5 independent of the number of processors
- Parallel interpolator for partitioned HDF5 meshes
- Automatic sponge layers
- Built-in Gaussian filters of arbitrary size
Software engineering
- 185,000 lines of code
- Object-oriented fortran with modules (f90)
- Version management with GIT
- Inline documentation in the source code (XML + Latex)
- GUI with client/server mode (wxwidgets, C++)
- Automatic validation tests (AQAT, AVVT)
- Automatic dependency of f90 modules in makefiles
- Keyword-based input file
- Easy profiling with timers
Gallery
Some computation examples are given in the gallery.
Performances
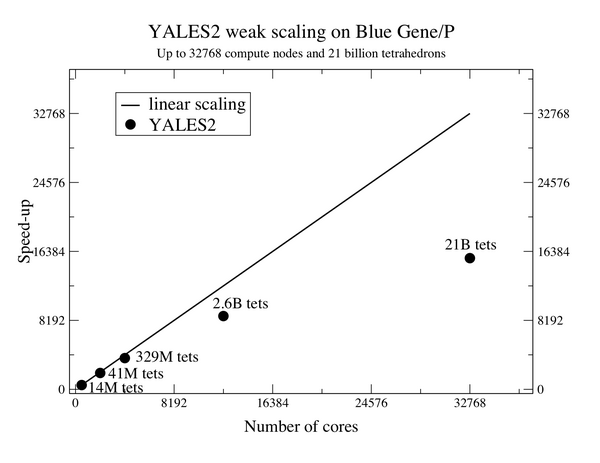
Thanks to highly efficient linear solvers, the speed-up of YALES2 is almost linear for meshes with several billion elements. These measures up to 21 billion elements were performed at IDRIS in France and at the Juelich Supercomputing Center in Germany.